| Authors: |
Christophe Laummonerie, Jerome Mutterer, Institut de Biologie Moleculaire des Plantes, Strasbourg, France. Philippe Carl (maintainer), UMR7021, Strasbourg, France. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| History: |
2004/02/02: First version: Parts of the initial code were taken from plugins by W. Rasband, P. Bourdoncle, and G. Chinga. 2005/01/28: Version 1.1: Statistical methods added. 2006/08/29: Version 1.2: - Ratio bars non destructively displayed - Works with any closed ROI - Fire LUT better diplayed - You now have to click in the ROI to update overaly image(s) and statistics 2016/03/21: Version 1.3: - Replacement of the deprecated functions (getBoundingRect, IJ.write) by the new ones - Extension of the plugin for whatever picture dynamics - Addition of a plot (with legends, ticks (minor and major), labels) within the scatter plot - The selected points within the overlay picture are updated as soon as the ROI in the scatter plot is modified or dragged over - Possibility to move the ROI position (within the scatter plot) from the mouse position within the overlay picture - Possibility to set ROIs with given colors with a mouse double click - Possibility to generate the x or y histogram with a Gaussian fit in order to extract the histogram maximum position by using the numeric pad 4/6 or 2/8 keys 2019/10/14: Version 1.4: - Addition of scripting possibilities illustrated within the macro examples that can be downloaded here and here. - The colocalization calculations are performed using double parameters instead of float 2019/12/15: Version 1.5: - Possibility to add a selection within the Composite picture to restric the analysis to a given selection - Addition of synchronized background thread for smoothly updating the calculations on the fly 2022/06/12: Version 1.6: - Possibility to choose the size of the scatter plot upon start of the plugin - Addition of a label panel at the bottom of the scatterPlot picture displaying the limits of the scatterPlot Roi selection (or other parameters upon selection) - Addition of a "Set" button at the bottom left of the scatterPlot picture allowing so set the limits of the scatterPlot graph and/or of the scatterPlot Roi and/or choosing the displayed parameters within the label panel at the bottom of the scatterPlot (the 'g' key gives the same features) - Addition of the Mander's coefficients (M1 & M2) calculation - The possibility to set ROIs with given colors with a mouse double click has been erased (due to the ImageJ 1.53c 26 June 2020 update) and replaced by a Ctrl + mouse click user action 2023/03/18: Version 1.7: - Possibility to choose the size of the scatter plot upon start of the plugin 2023/05/01: Version 1.8: - The Colocalization_Finder plugin allows the analysis of image stacks 2024/06/20: - Addition of macros for illustrating the questions within the How to paragraph 2025/03/23: Version 1.9: - Addition of the linear fit line of the nb_pixels pixels selected within the scatterPlot ROI |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Source: | Contained in the JAR file. To open a JAR file, change the extension from ".jar" to ".zip" and double click on it.
or Download Colocalization_Finder.java. |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Installation: | Download Colocalization_Finder.jar to the plugins folder, or subfolder, restart ImageJ, and there will the "Colocalization Finder..." new command in the Plugins menu. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
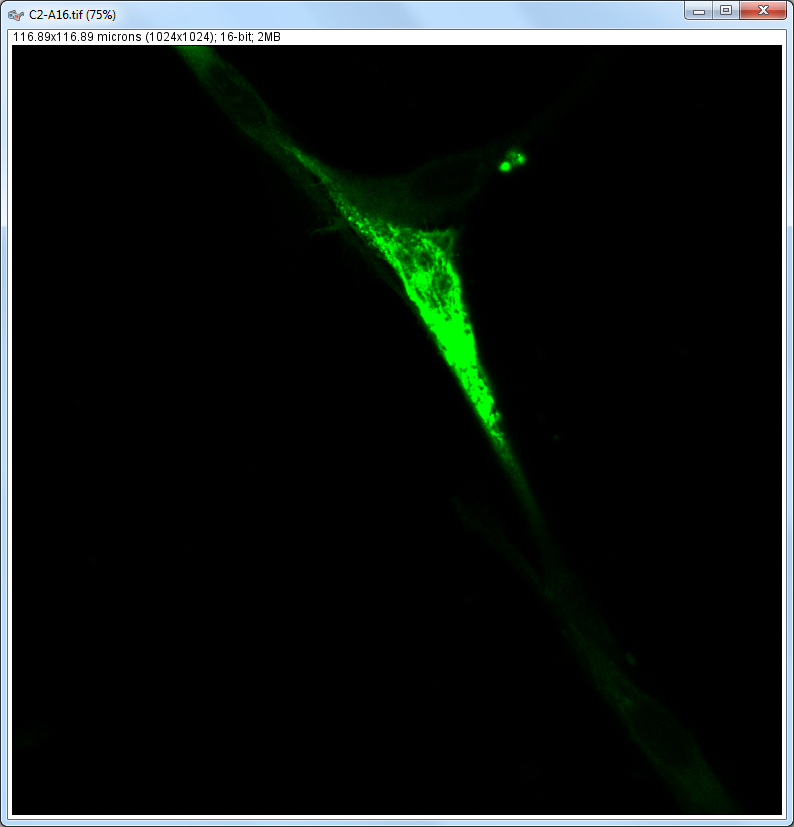
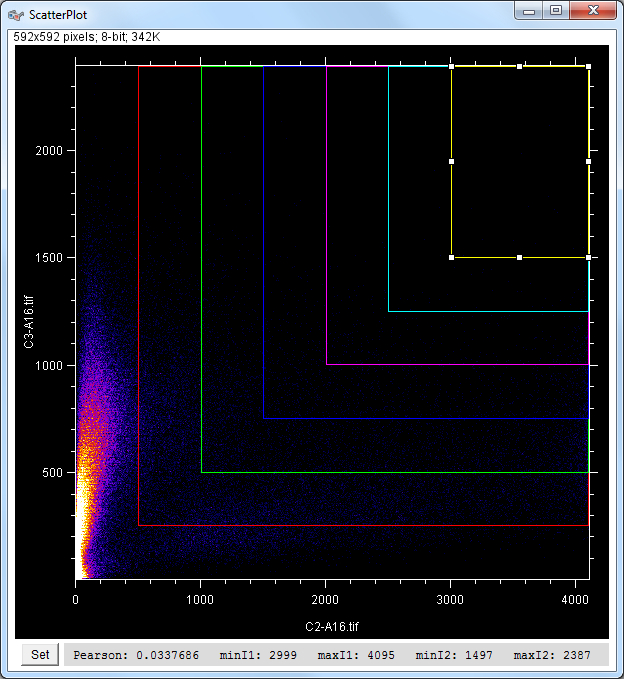
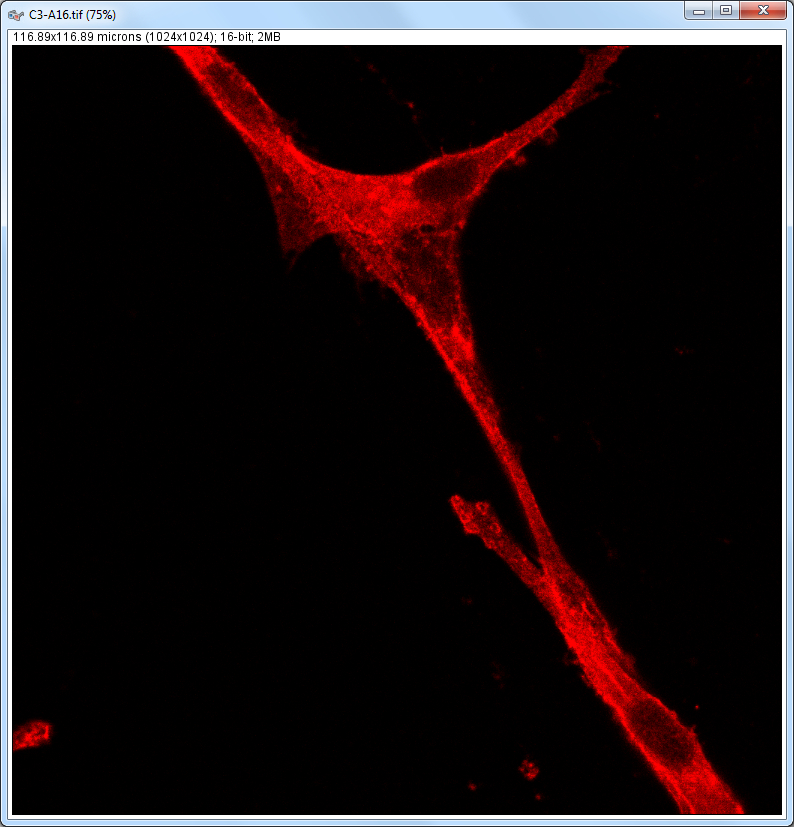
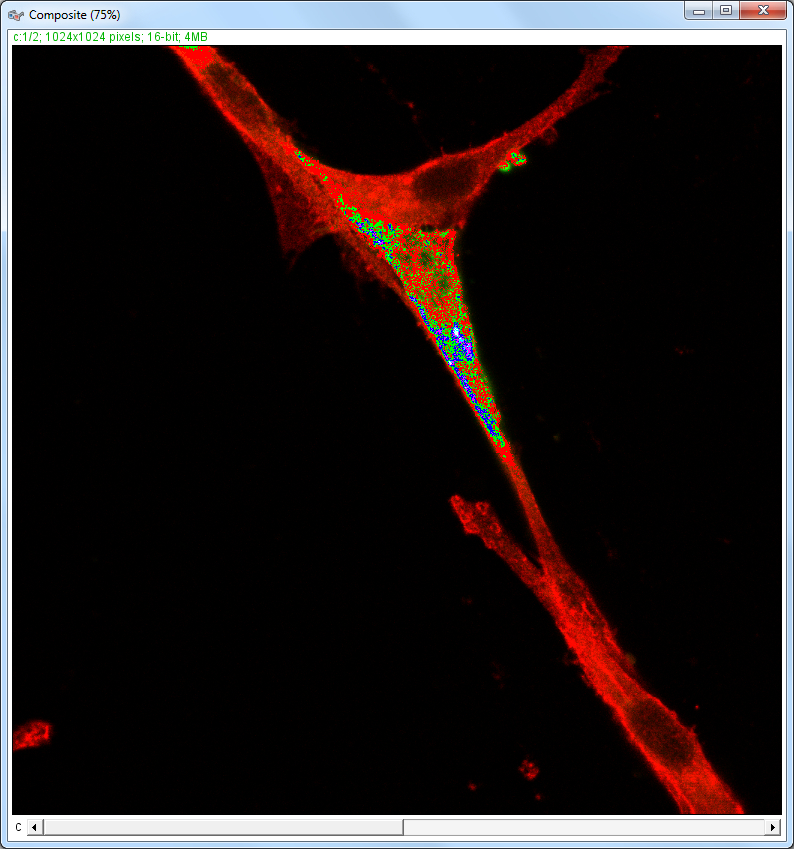
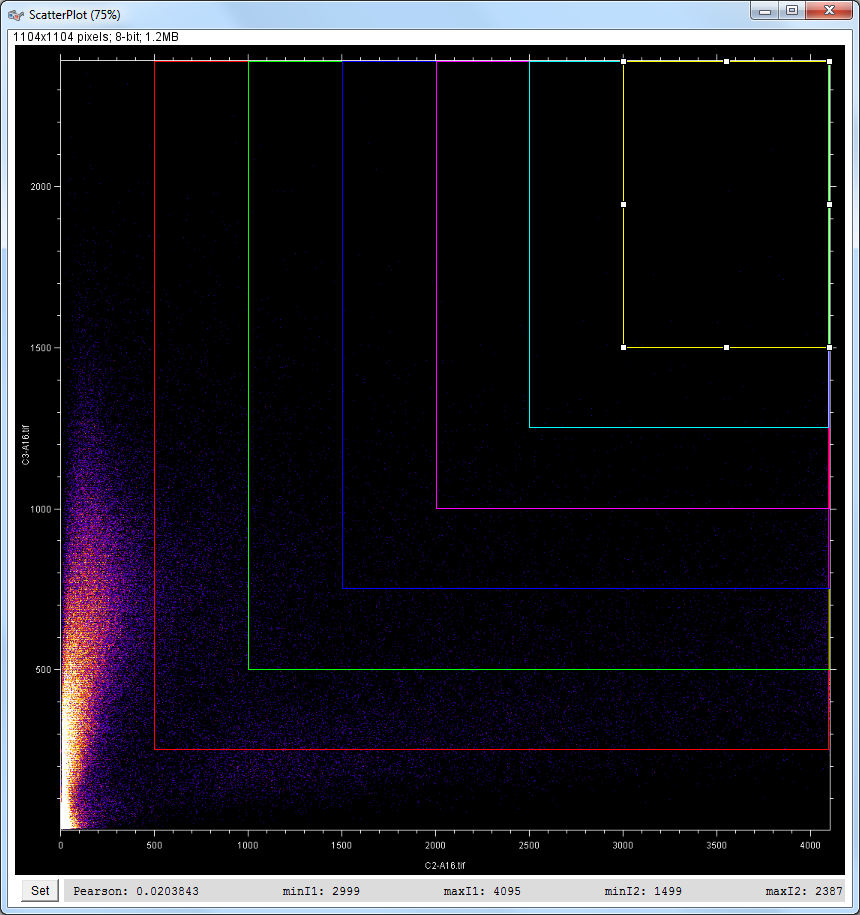
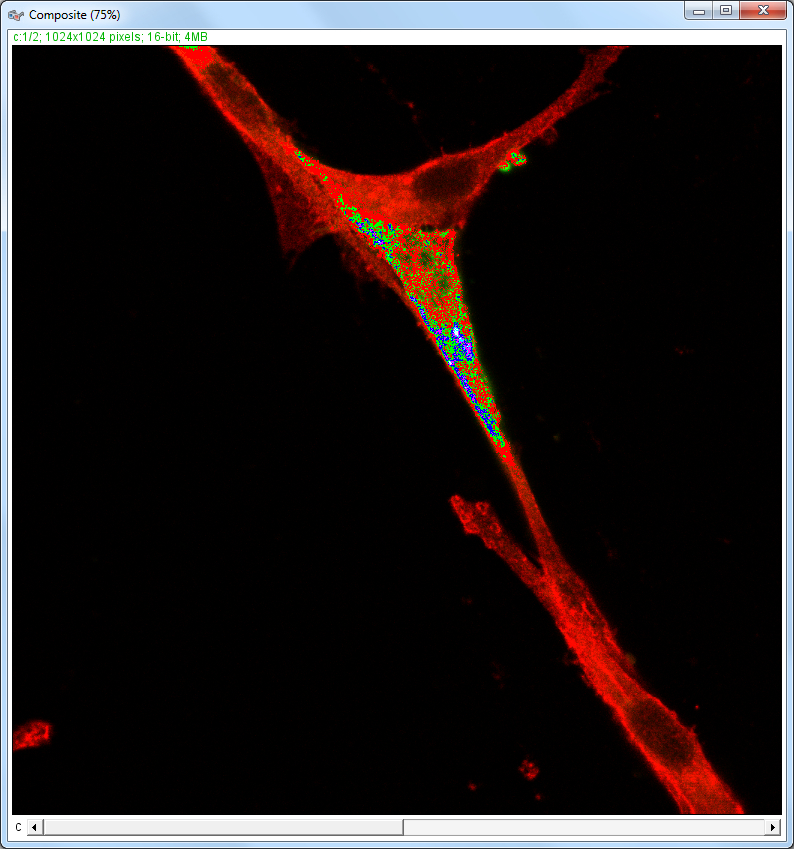
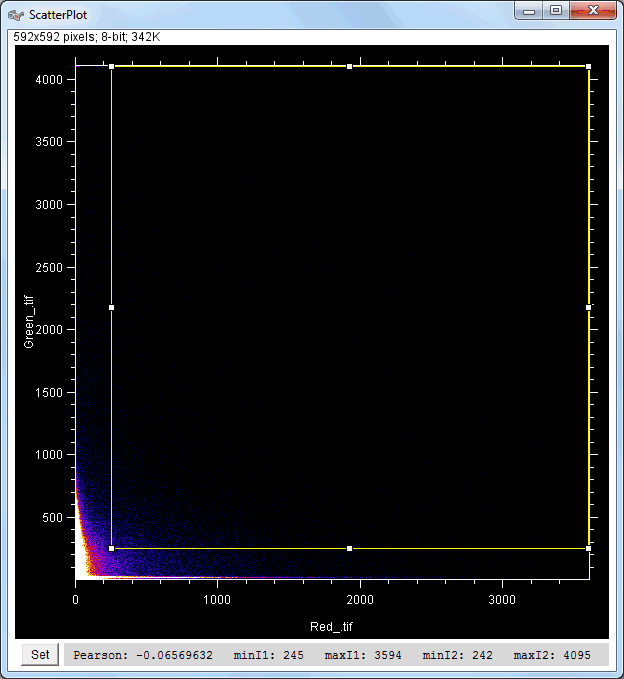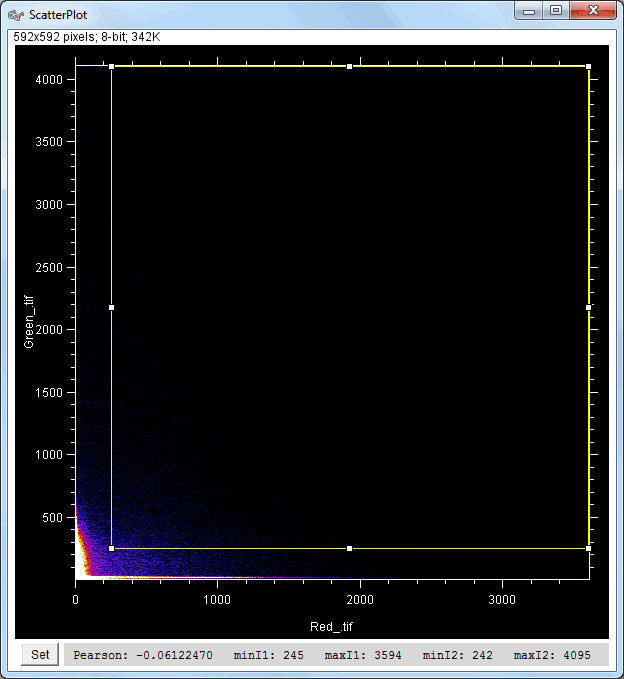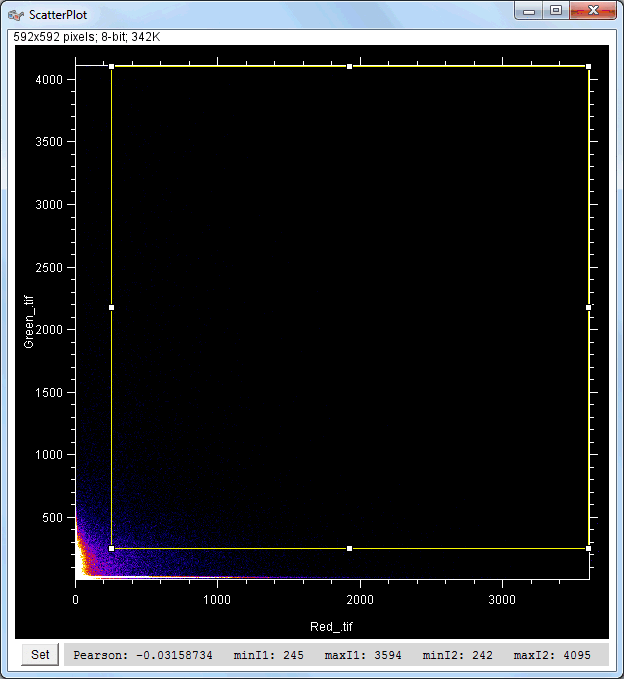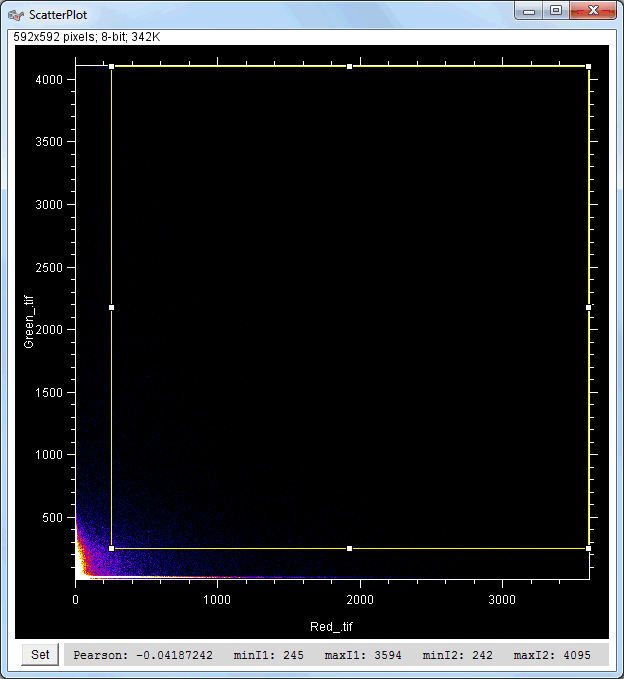
| Description and features: | This plugin displays a correlation diagram (called scatterPlot picture) from two initial pictures having the same size together with a RGB overlap of the original images (called Composite picture). The initial scatterPlot plot limits correspond to the minimal and maximal intensity values of the input pictures. The user interface of the plugin provides the following features:
Plugin launching interface: 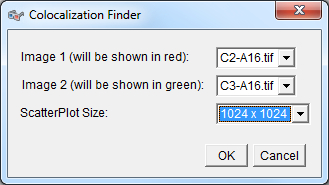
Example results (for a 512 × 512 scatterPlot):
Example results (for a 1024 × 1024 scatterPlot):
The results table with computer Pearson's correlation, Overlap coefficient, contribution of both channels to the overlap coefficient, Slope and intercept of the linear regression,% pixels selected, min and max in both channels. with \(\bar a\) and \(\bar b\) being the average values of a and b, i.e. \( \large { \bar a = { 1 \over n} \sum_\limits{i=1}^{n} a_i } \) and \( \large { \bar b = { 1 \over n} \sum_\limits{i=1}^{n} b_i } \) as \(\sigma_a\) and \(\sigma_b\) being the standard deviation of a and b, i.e. \( \large { \sigma_a = \sqrt { { 1 \over n-1 } \sum_\limits{i=1}^{n} ( a_i - \bar a )^2 } } \) and \( \large { \sigma_b = \sqrt { { 1 \over n-1 } \sum_\limits{i=1}^{n} ( b_i - \bar b )^2 } } \) which gives: Pearson's_Rr \( \Large { = { 1 \over n-1 } \sum_\limits{i=1}^{n} { a_i \; - \; \bar a \over \sqrt { { 1 \over n-1 } \sum_\limits{i=1}^{n} ( a_i \; - \; \bar a )^2 } } \times { b_i \; - \; \bar b \over \sqrt { { 1 \over n-1 } \sum_\limits{i=1}^{n} ( b_i \; - \; \bar b )^2 } } } \) \( \Large { = { 1 \over n-1 } \sum_\limits{i=1}^{n} { { (a_i \; - \; \bar a) \; \times \; \sqrt { n-1 } } \over \sqrt { \sum_\limits{i=1}^{n} ( a_i \; - \; \bar a )^2 } } \;\;\, \times { { (b_i \; - \; \bar b) \; \times \; \sqrt { n-1 } } \over \sqrt { \sum_\limits{i=1}^{n} ( b_i \; - \; \bar b )^2 } } } \) \( \Large { = { 1 \over n-1 } \sum_\limits{i=1}^{n} { { \sqrt { n-1 } \; \times \; \sqrt { n-1 } \; \times \; (a_i \; - \; \bar a) \; \times \; (b_i \; - \; \bar b) } \over { \sqrt { \sum_\limits{i=1}^{n} ( a_i \; - \; \bar a )^2 } \; \times \; \sqrt { \sum_\limits{i=1}^{n} ( b_i \; - \; \bar b )^2 } } } } \) \( \Large { = { n-1 \over n-1 } \quad\;\,\, { { \sum_\limits{i=1}^{n} ( a_i \; - \; \bar a ) \; \times \; ( b_i \; - \; \bar b ) } \over { \sqrt { { \sum_\limits{i=1}^{n} ( a_i \; - \; \bar a )^2 } \; \times \; { \sum_\limits{i=1}^{n} ( b_i \; - \; \bar b )^2 } } } } } \)
M1 \( \Large = { \sum_\limits{i=1}^{n} a_{i,coloc} \over { \sum_\limits{i=1}^{n} a_i } } \) with \( \begin{cases} a_{i,coloc} = a_i & \text{if } b_i > b_{treshold} \\ \\ a_{i,coloc} = 0 & \text{if } b_i \leqslant b_{treshold} \\ \end{cases} \)
The Colocalization_Finder plugin version details as well as can be reached from the ImageJ Help>About_plugins>Colocalization_Finder...Help>About_plugins>Colocalization_Finder...: 
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
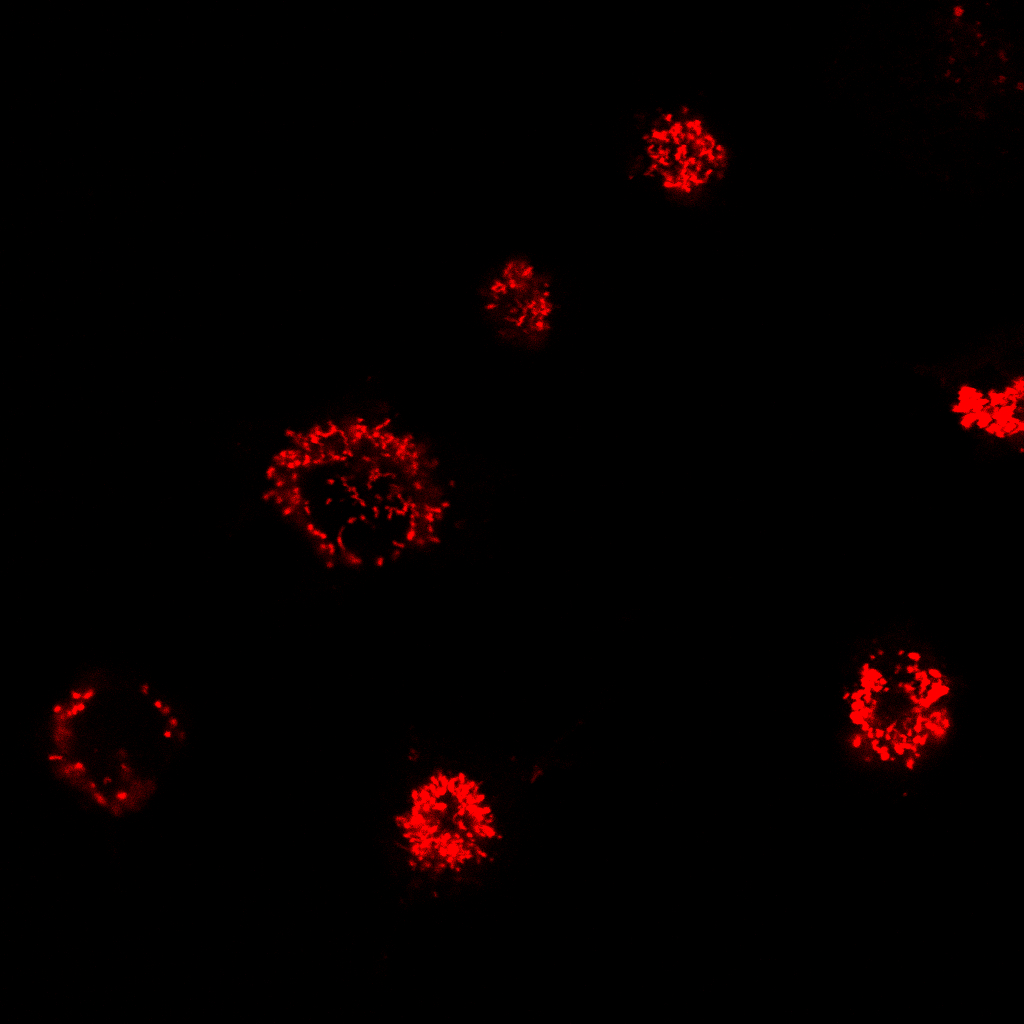
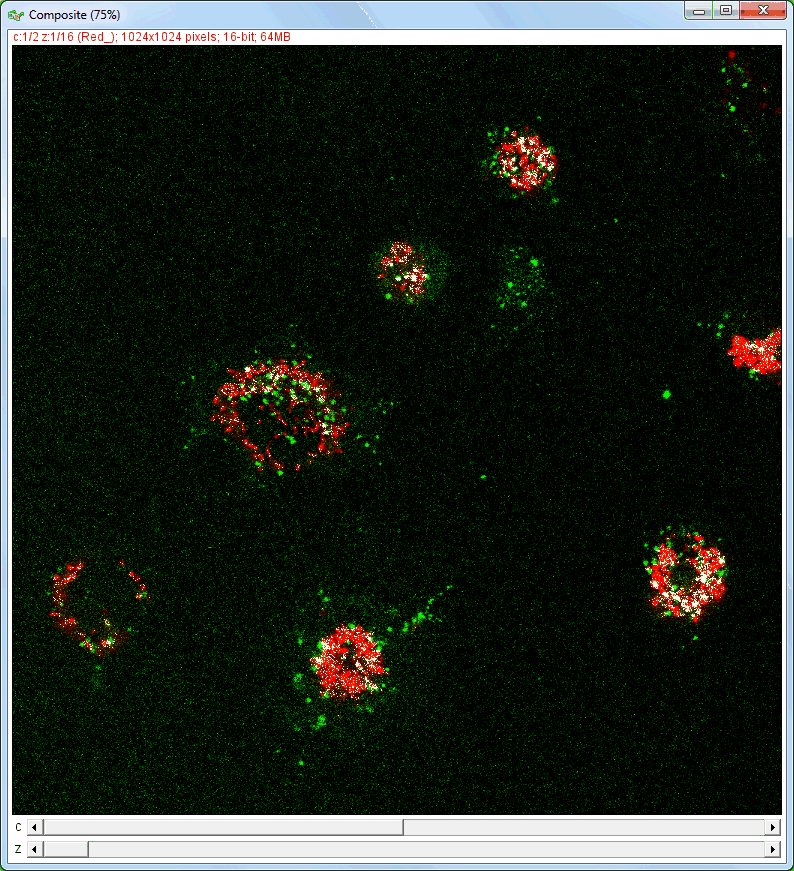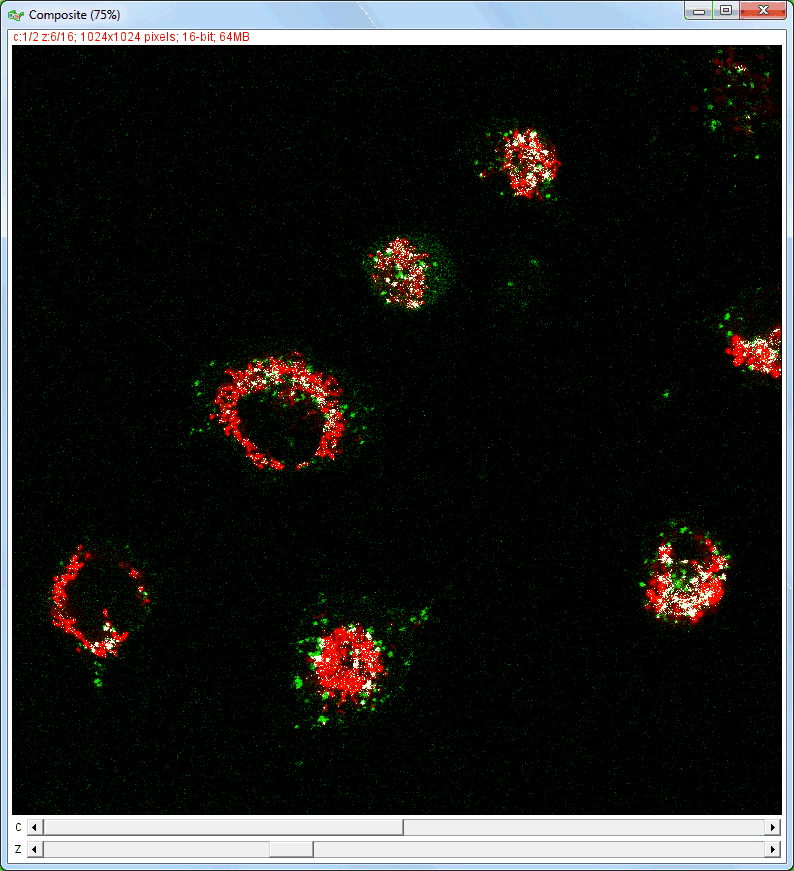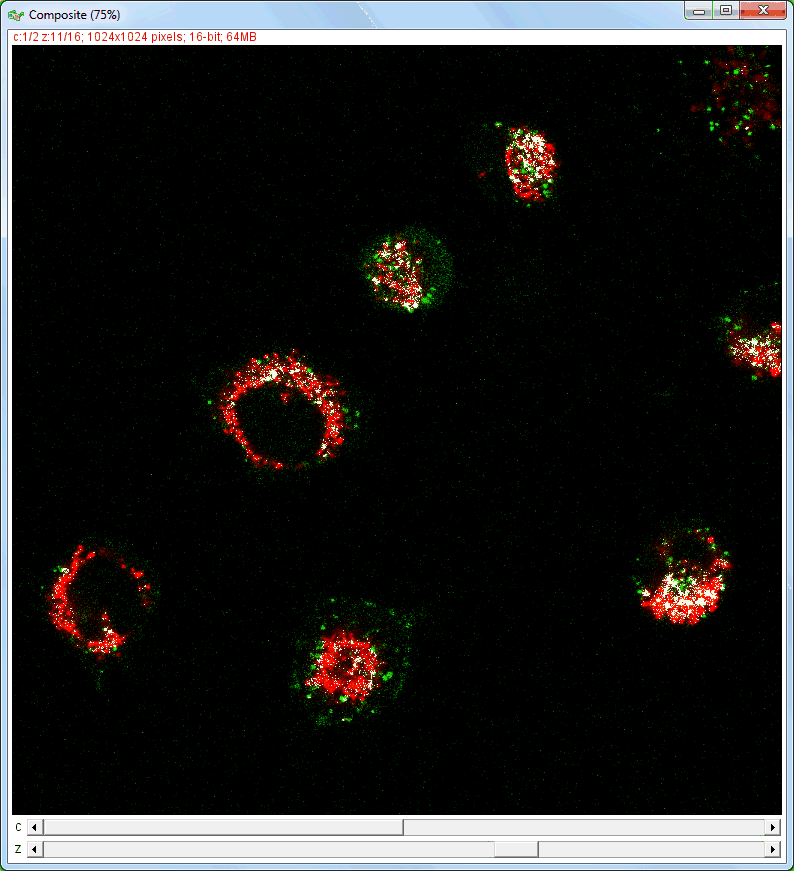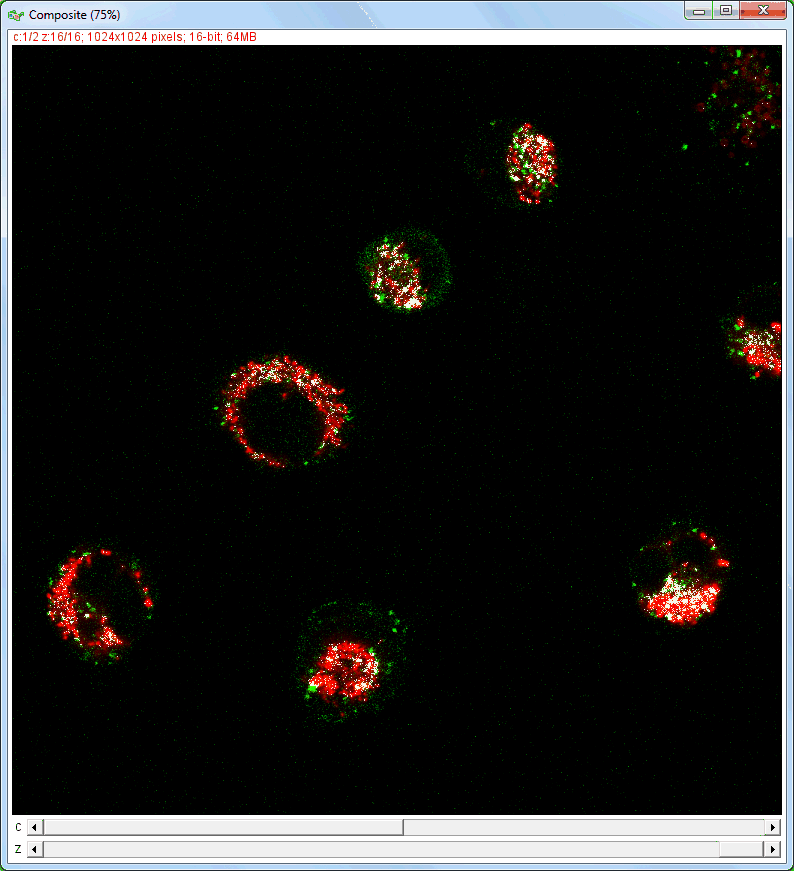
| Analysis of image stacks: |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Macro functions: |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| How to: |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Validity verification: |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Webinar: |
A webinar on how to use the plugin can be seen here: |
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||